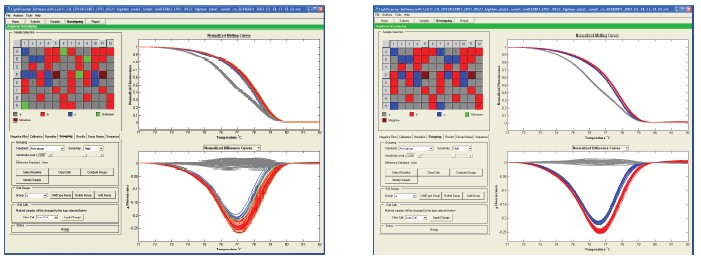
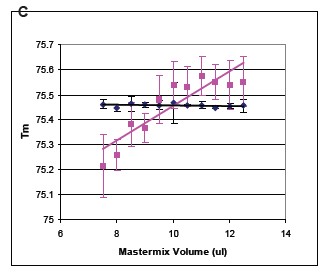
Summary Genetic typing using high-resolution melting curve techniques is not only simple but effective. In the analysis method based on the melting curve, the probe method is a gold standard for genotyping, and can comprehensively detect the matched target allele. Small fragment amplification is a method that does not require the design of a probe for genotyping. Due to the presence of heteroduplexes, heterozygotes are readily detected, which largely alters the peak shape of the melt-transformed. Changes in G/C or A/T in homozygous genotyping are the most difficult to detect because their alleles have similar Tm values. Base pairs called neutral homozygotes are difficult to distinguish using the small fragment method. In addition, homozygous indistinguishable is also limited by changes in temperature between the sample and the sample, buffer and volume. Regardless of the precise instrumentation, the relevant changes may be significant, so a method is needed to reduce this variation and improve small fragment genotyping. These limitations can be overcome by using specific internal standards. background Synthetic oligonucleotide internal and complementary sequences contain the same molecular concentration. In the presence of an internal standard, a conventional 96-well plate PCR machine was used, and LC Green® Plus dye was added for PCR reaction. After the completion of the PCR reaction, the PCR product was placed in a Lightsanner instrument, and melting curve analysis was performed at 55 ° C to 97 ° C. The internal standard's dissolution identification is calibrated by the fluorescence data of each sample. The genotype was verified by sequencing. The error rate of homozygous genotype detection before and after correction was estimated by measuring the distance between the peaks of the Tm values ​​of the corresponding genotypes of each sample group. When the Tm value of the sample is close to the Tm value of the error group, an error will be detected. result By using different microplates and PCR template validation, the sensitivity of pure and genotypes to neutral single nucleotide polymorphism base pair detection was increased by 90% to 99% after calibration with temperature internal standards. The Tm value is lower by about 0.056 to 0.027 ° C after using the temperature internal standard. in conclusion The internal standard increases the changes in pure and genotype G/C and T/A bases within small fragments. The study was supported by the following sub-sectors of the Ministry of Health: R44DK069106, R44HD053215 to SFD, R42GM072419 and R42GM073396 to CTW. background When denatured or melted, the genotype signal changes due to the inherent properties of the nucleic acid. Probe-based genotyping has a large advantage in the large number of related allele specificities in temperature changes. The high resolution melting curve provides a melting map of the entire double strand and the probe can be removed. In addition, the number of base interrogations obtained increased by a factor of 20 per experiment when the entire amplicon was monitored. Due to the very similar melting map and melting temperature, it is difficult to detect pure and genotypes without probes. The small fragment provides a simple genotyping program. method 10 μL of the PCR reaction system was added with 1X LightScanner High Sensitivity Mastermix, which contains hot start enzyme, magnesium ion buffer, other components and nucleic acid calibration. The PCR melting range is about 62C and 92C. Primers of 0.10 μM or 0.15 μM were included in the system, and human genomic DNA was 10-15 ng. The PCR cycling conditions were as follows: 95 C pre-denaturation for 2 min, 94  C/30 s, 64-67  C/30 s (depending on the experiment) including annealing and extension, 45 cycles. The final annealing condition was 28 C/30 s. The genotype of all DNA samples was detected by probe method. result The complete melting diagram before using the temperature internal standard is shown in Figure 1. The melting diagram from the low temperature internal standard to the high temperature internal standard is indicated, and the middle is the melting diagram of the amplicon. Figure 1. The derived melting curve shows the internal standard and the melting peak of the amplicon. The melting peak of the internal standard is on the left and right sides. 94 melting plots were generated from independent PCR reactions with melting peaks appearing at approximately 76 °C. The internal standard molecules were not calibrated and the homozygous peaks were not completely separated. The narrowest and highest peaks in the middle are A/A (blue) and T/T (red), which are pure and genotypes of the CPS1 A/T polymorphism. Previous probe-based genotyping colors have increased, not based on this small fragment melting data. The other peak in the middle is the A/T heterozygous peak. The inset shows the amplified melting peak. Figure 2. Using a temperature internal standard can improve the melting curve of the derivatization. Fluorescence plots show 94 independent PCR reactions for CPS1, OTC, MSH2 and PAH mutations. Bimodal CPS1 heterozygotes are easily distinguished before calibration (Figure 2a). In contrast, OTC heterozygotes (Fig. 2c) show only one major peak, and more peaks are difficult to distinguish before calibration. It is easier to use the genotype data after the temperature internal standard, and no different genotypes are found in the repeated reactions. Table 1 compares the four replicate Tm before and after the temperature internal standard for each gene of interest. In all cases, the difference after calibration is reduced. It can be seen that the difference between the standard deviation and the non-interest area after calibration is reduced.                                                     Table 1. Statistics of the impact of calibration on Tm values Table 1. Each replicate disk contained 47 genomic DNA samples of the same setup (94 independent PCR reactions). Tm predicted based on all homozygous neighbor parameters is listed in the table. Heterozygous Tm values ​​are not included as they are usually typed in our system without calibration. To some extent, due to the presence of LCGreen dye, the actual Tm value obtained by stable DNA hybridization is 5-7 ° C higher than the theoretical value. Genotyping without genotyping is a significant difference between uncalibrated and calibrated Tm values. (p<0.001). Table 2. Calibration improves homozygote detection sensitivity Table 2. Experimental data shows the effect of calibration on sensitivity. It can be seen that the detection sensitivity after calibration is increased by 10%. In the experiment, the correct estimate is based on the approximate Tm value, not the Lightsanner software prediction. A miscalculated prediction of each target small segment will be displayed. Software automatic calibration Subsequent work was to automatically group samples of similar images using Lightsanner software. Genotyping of this pattern does not require knowledge of the Tm value. This type of graph-based typing has benefited from the use of temperature internal standards for genotyping based on this Tm value. Figure 3 shows that the improvement by calibrating the small fragment product of CPS1. Figure 3. The temperature internal standard improves the Autosanner software auto-calibration. The lower panel shows the normalized melting curve and different screenshots. In addition, as can be seen from the grouping situation, the red-blue ash color of the box on the left side of the screen is a repeating pattern in columns 1-6 and 7-12. The sensitivity obtained before calibration of this data set was 92%, and the sensitivity obtained after calibration was 100%. Calibration reduces the dependence of Tm on the amount of reaction The Tm value is predicted by the amount of reaction because more Mastermix or mineral oil in the reaction plate will hinder the transfer of heat. This will increase the displayed Tm value. We used OTC small fragments to test the dependence of Tm values ​​on the volume of the Mastermix reaction. From 7.0 μL to 12.5 μL in 0.5 μL increments (10 μL recommended by the manufacturer). The dependence of the Tm value on the amount of reaction was large before calibration (slop = +0.069, R2 = 0.078), but after calibration (slope = -0.001, R2 = 0.066), see Figure 4. Calibration can effectively reduce the effect of the amount on the Tm value. Figure 4. Effect of Mastermix volume on Tm values ​​before and after calibration. 47 samples of the amplified OTC gene were evaluated for the C.299-8T>A mutation. The reaction was supplemented with 1 μL of DNA template using 7.5 1 μL to 12.5 1 μL of Mastermix in 0.5 μL increments. Panel A shows a melting map of important mutations prior to calibration. Panel B shows the reduced mutations after calibration. The C plate shows the amount dependence of the Tm value on the Mastermix before the calibration of the 35T/T homozygous sample. Two conditions can be observed after calibration: 1) the error line will be compressed, the mutation display will be reduced 2) the slope and R2 value will be greatly reduced, indicating that the Tm value has little to do with the amount of Mastermix. The distribution of the different orientations of each reaction in the plate is to reduce the dependence of the Tm value on the position of the sample. in conclusion Small fragment melting is a simple genotyping method that does not require post-PCR processing of the sample. Although smaller fragments are well qualified for genotyping. However, the success of homozygous genetic analysis without temperature internal standards depends not only on high-resolution data collection, but also on the uniformity of buffer, volume, and extraction between samples. Efficient calibration can eliminate these changes in mixing factors for better, relatively stable homozygous genotyping. references 2. Nellaker, C., Wallgren, U., Karlsson, H. (2007) Molecular beacon-2. based temperature control and automated analyses for improved resolution of melting temperature analysis using SYBR I green chemistry. Clin Chem. 53, 98- 103. 3 .Liew, M., Seipp, M., Durtschi, J., Margraf, RL, Dames, S., 3. Erali, M., Voelkerding, K., Wittwer, C. (2007) Closed-tube SNP genotyping Without labeled probes/a comparison between unlabeled probe and amplicon melting. Am. J. Clin. Pathol. 127, 341-348 4. Seipp, MT, Durtschi, JD, Liew, MA, Williams, J., 4. Damjanovich, K., Pont-Kingdon, G., Lyon, E., Voelkerding, KV, Wittwer, CT (2007) Unlabeled Oligonucleotides As Internal Temperature Controls for Genotyping by Amplicon Melting. J Mol Diagn., 9, 284-9. Surgical Tape,Adhesive Flexible Tape,Waterproof Surgical Tape,Breathable Surgicaltape Zhende Medical Co.,Ltd , https://www.zdmedicalproduct.com
It is worth noting that small fragment amplification is better for homozygous detection than large fragment amplicon because the specific genotype Tm value will change more. However, certain factors can affect the separation of large numbers of homozygotes. Genotyping can vary from base type Tm values ​​to device changes and sample and sample differences. Temperature internal standards can reduce these changes, fix the melting transition and increase the sensitivity of genotyping. The best results will be obtained when the small fragment primers match only the flanking SNPs.
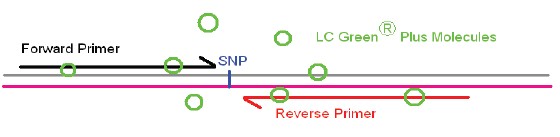
Melting was performed on a 96-well plate LightScanner device with data acquisition from 55-97 ï‚°C. The data calibration after melting first uses a smooth line close to the fluorescence data, and then the Tm value is calculated using the line interpolation peak of the data. This adjustment process calibrates the melting peaks of each amplicon to achieve minimal changes. The data was re-analyzed using LightScanner software. The temperature region near the amplicon Tm value was used for analysis (with or without previous internal standard calibration) while being shown as a derivative peak.


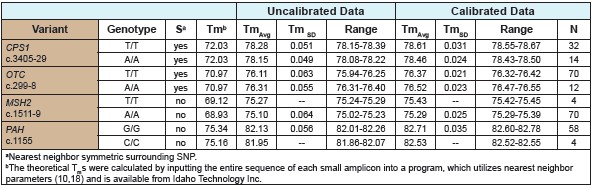
For the CPS1 and OTC PCR products, no difference in homozygous T/T and A/ATm was found by analysis of the predicted neighbor values. However, the two target genes, even if not calibrated, have different Tm values ​​for the homozygous population. Although, from the range of Tm values ​​seen, the homozygous T/T and A/A overlap before calibration. After calibration, the homozygous T/T and A/A did not overlap, indicating that homozygotes have been distinguished (Table 1). For MSH2 and PAH small fragment products, proximity analysis predicts a small difference in Tm values ​​between pure and amplicon. Analysis of homozygous changes before calibration showed that genotypes could not be clearly distinguished. After calibration, the error estimates for homozygous genotypes are shown in Table 2.
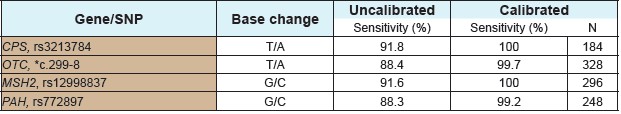
* The number of rs of the OTC T/A mutation is not indicated.

1. Dodge, A., Turcatti, G., Lawrence, I., de Rooij, NF, Verpoorte, 1. E., (2004) A microfluidic platform using molecular beacon-based temperature calibration for thermal dehybridization of surface-bound DNA Anal Chem. 76, 1778-1787.